BioInformatics
   NGS data interpretation and the ability to re-mine that data is increasingly important to companies. SeqLL provides a complete bioinformatics solution to help maintain timelines and identify critical knowledge related to drug candidate action and effect. Our implementation for each project is specific, efficient and cost effective. SeqLL bioinformatics specialists will work with your team to define and confirm your exact requirements and deliver information that answers the questions your program is investigating. Once complete, the results are reviewed with you at a closing meeting or the next step in the program. NGS data interpretation and the ability to re-mine that data is increasingly important to companies. SeqLL provides a complete bioinformatics solution to help maintain timelines and identify critical knowledge related to drug candidate action and effect. Our implementation for each project is specific, efficient and cost effective. SeqLL bioinformatics specialists will work with your team to define and confirm your exact requirements and deliver information that answers the questions your program is investigating. Once complete, the results are reviewed with you at a closing meeting or the next step in the program.
Interpretation - including raw reads, filtered reads, aligned reads (when applicable), sequence information and data files are provided with each sequencing project.
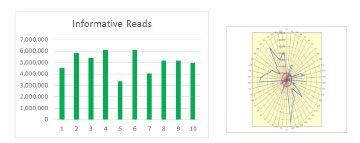
Basic Bioinformatics - Data interpretation starts with informed reads, quality control, Digital Gene Expression (DGE), box plots and bar graphs.
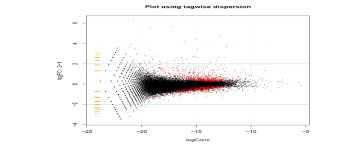
Advanced Bioinformatics - Typically this expands the report to include Differentially Expressed Gene (DEG), gene ontology and pathway analysis to gain additional insight.
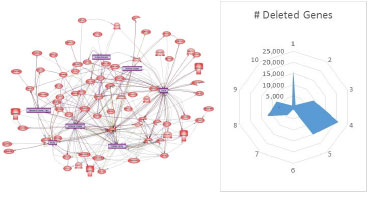
Custom Bioinformatics - combine your molecule or target specific expertise with our analysis experience to gain non-intuitive insight to ADARediting, vlincRNA construction, CAGE analysis and other specific complex statistical analysis. Re-evaluate existing data for additional correlation and discovery.
|