BioInformatics
Innovative sequencing approaches are generating vast amounts of data. Turning raw data into real insights has never been more important or challenging. Bioinformatics experts at SeqLL offer a range of services to help researchers reach their goals.
Read More... |
|
DRS - Direct RNA sequencing For the first time, the RNA transcriptome can be explored directly as it exists in-vivo, with no conversion to cDNA required. SeqLL has pioneered Direct RNA Sequencing (DRS) and is now offering DRS services to the research community.
Read More... |
|
FFPE (Formalin Fixed Paraffin Embedded) Samples SeqLL's sequencing technology works best on short fragments - as low as 50 base pairs – so it is ideal for degraded, poor quality samples like FFPE. No additional kits, amplification or work-arounds are needed.
Read More... |
|
|
Epigenetics
The ability to detect, quantify, and combinatorially map modified nucleosomes is critical to deepening our understanding of epigenetics. SeqLL’s tSMS is the only platform in which researchers can correlate modifications on the genome with sequence data at a single molecule level.
Read More...
|
|
|
 SeqLL's Sequencing Lab is Now Processing a Wide Range of Samples SeqLL's Sequencing Lab is Now Processing a Wide Range of Samples
 tSMS provides high accuracy and sensitivity to investigate areas that other NGS platforms avoid or miss. Generate more unique reads and greater depth of coverage without amplification, no library prep and no bias from heterogenous DNA or RNA samples. tSMS provides high accuracy and sensitivity to investigate areas that other NGS platforms avoid or miss. Generate more unique reads and greater depth of coverage without amplification, no library prep and no bias from heterogenous DNA or RNA samples.
The tSMS provides:
- High Accuracy Sequencing
- Simple and efficient sample preparation process
- No libraries to prepare
- Bias free results – Amplification not required
- Identification of low abundance transcripts
- High Precision – correlation between runs meets or exceeds R > 0.99
Flow cell surface chemistry is optimized for rapid hybridization and dense packing of genetic material from each sample while allowing signal from each strand to be individually captured and recorded. Multiplexing and high sample throughput are possible. The platform reads each strand of DNA and RNA by synthesis to ensure accuracy. Direct RNA sequencing, sheared, fragmented or short RNA species, CNV, indels, chromosomal abnormalities, and differential gene expression analysis are easily performed with tSMS technology.

|
|
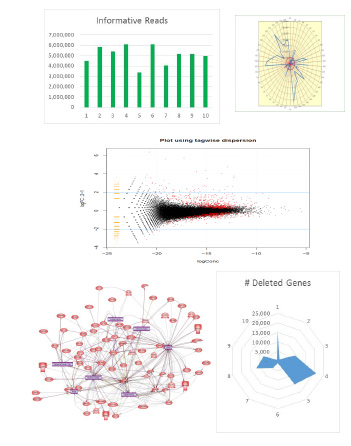 Sophisticated NGS data interpretation is increasingly important to companies. SeqLL provides a complete bioinformatics solution to help you meet your deadlines and make critical discoveries related to drug candidate action and effect. Our implementation for each project is specific, efficient and cost effective. SeqLL bioinformatics specialists will work with your team to define and confirm your project requirements and to deliver a custom data analysis package that meets the specific needs of your program. Once complete, the results are reviewed with you as a closing or next step in the process.
Sophisticated NGS data interpretation is increasingly important to companies. SeqLL provides a complete bioinformatics solution to help you meet your deadlines and make critical discoveries related to drug candidate action and effect. Our implementation for each project is specific, efficient and cost effective. SeqLL bioinformatics specialists will work with your team to define and confirm your project requirements and to deliver a custom data analysis package that meets the specific needs of your program. Once complete, the results are reviewed with you as a closing or next step in the process.
Sequencing Metrics - including raw reads, filtered reads, aligned reads (where applicable) sequence information and data files are provided with each sequencing project.
Basic Bioinformatics - Basic data interpretation includes informed reads, quality control, box plots and bar graphs. Our data presentations are designed to enable intuitive visual inspection of the data.
Advanced Bioinformatics - Complex analyses provide additional insights and includes basic bioinformatics plus Differentially Expressed Gene (DEG), gene ontology and pathway analyses
Custom Bioinformatics - combine your molecule or target specific expertise with our analysis experience to gain non-intuitive insight into your data using our team’s expertise in complex statistical analyses. Re-evaluate existing data for additional correlation and enhanced discovery.

|
|
DRS™ – Direct RNA Sequencing
Only SeqLL offers Direct RNA Sequencing without conversion to cDNA. The technology offers direct observation of RNA and allows a more complex and accurate understanding of the transcriptome without sample prep bias or library artifacts. As with all of our true Single Molecule Sequencing services, our amplification-free platform enables accurate quantification of transcripts and unrivaled sensitivity to low-fold changes of gene expression.
DRS
- Simplified Sample Prep
- Single molecule synthesis and reads
- Bias free data
- Authentic transcriptome detail
- Evaluate 3' end of transcripts for gene expression studies
- Detect low fold changes in expression to uncover novel biomarkers

|
|
FFPE – Formalin Fixed Paraffin Embedded Samples
FFPE samples often exhibit degradation during processing or storage that can challenge other NGS systems to provide meaningful information, as these systems require approximately 500 base pairs for analysis. Kits and work-arounds have been developed to enable analysis of FFPE samples with standard NGS, however, they utilize further amplification that can introduce significant bias. SeqLL's tSMS technology is uniquely suited to sequence degraded DNA and RNA fragments from FFPE tissue due to its short read capability and amplification-free methodology. As a result, no additional bias is introduced and no additional kits or procedures are required.

|
|
cDNA based – RNA Sequencing
SeqLL’s quantitative sequencing platform allows investigators to design gene expression research across oncology, infectious diseases, age-related disorders and genetic diseases. The approach is unique in its ability to identify low-fold changes and discover rare transcripts that are missed by other sequencing methods.
Typical applications include:
- Study of the effect of transcriptome or phenotypes associated with disease progression
- Assessment of gene expression coincident with responses to standard and investigational treatment regimens
- Identification of both dominant and rare species
- Evaluation of the function of low abundance, non-coding transcripts
- Splice Variant Analysis to assess the expression rate of the exons across a transcript
- Observe low fold changes to expression as part of drug development and surveillance programs
- Biomarker identification via profiling subtle changes in transcript levels for normal vs control subjects.

|
|
Application Highlight
Epigenetics - is the study of environmental stresses creating changes in gene expression that can be passed to offspring. Theses changes leave the underlying DNA sequence untouched but affect gene expression (up or down), resulting in a new phenotype. These environmentally induced changes can also be transient (and potentially) reversible - creating an extremely complex situation to study and understand. By better understanding how this process works, researchers hope one day to be able to use these mechanisms to selectively switch genes on or off. This is a new and exciting area of drug development that is being applied to many important disease areas.
 The Collaboration - SeqLL realizes that academic collaborations are critical to advancing our technology platform and to developing cutting edge applications that utilize our unique capabilities. Dan Jones, SeqLL's founder and President, worked closely with long-time collaborators Brad Bernstein, MD, PhD, and Efrat Shema, PhD to adapt SeqLL technology to examine correlations between histone modifications and sequence of the associated DNA. This novel approach promises to open a new frontier of single molecule epigenetics, enabling researchers to answer questions about epigenetic control of gene expression at a level never before possible. The Collaboration - SeqLL realizes that academic collaborations are critical to advancing our technology platform and to developing cutting edge applications that utilize our unique capabilities. Dan Jones, SeqLL's founder and President, worked closely with long-time collaborators Brad Bernstein, MD, PhD, and Efrat Shema, PhD to adapt SeqLL technology to examine correlations between histone modifications and sequence of the associated DNA. This novel approach promises to open a new frontier of single molecule epigenetics, enabling researchers to answer questions about epigenetic control of gene expression at a level never before possible.


|
|
|
|