|
 SeqLL’s Sequencing Services Laboratory offers a full range of bioinformatics services from Basic to Advanced to Custom analyses. SeqLL provides basic bioinformatics analysis as part of its standard sequencing service and also offers a menu of stand-alone bioinformatics services for data generated at other sites, including a variety of sequencing platforms.
SeqLL’s Sequencing Services Laboratory offers a full range of bioinformatics services from Basic to Advanced to Custom analyses. SeqLL provides basic bioinformatics analysis as part of its standard sequencing service and also offers a menu of stand-alone bioinformatics services for data generated at other sites, including a variety of sequencing platforms.
We recognize that the value of your NGS data is dependent on the quality of the analysis. Increasingly complex analysis is becoming routine as researchers strive to leverage the most they can from sequencing runs. Combining these issues with shorter and shorter timelines has raised unique issues for many research labs.
SeqLL offers a wide variety of bioinformatic packages, allowing the researcher to customize the analysis to fit the needs of a given project. SeqLL bioinformatic specialists will work with your team to define your project requirements and develop an analysis package that meets the specific needs of your program. Balancing cost, timing and level of analysis assures that you will receive a cost effective program, on time, and with the details that you require.
Sequencing Metrics are evaluated and delivered with the raw data in every project. Users select the file format that best suits their needs. This includes quality scores, evaluation of raw and filtered reads and where applicable aligned reads.
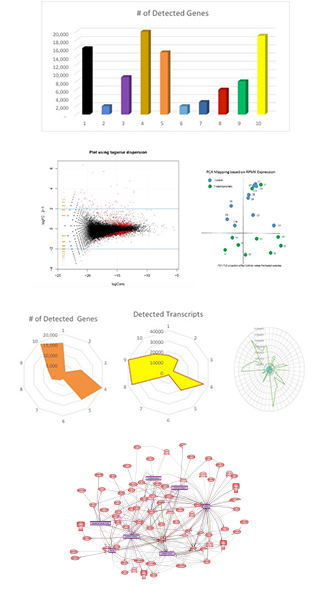
Basic Biometric interpretation is designed to provide intuitive visual evaluation of the data presented. The format will vary depending on the needs of the laboratory but will typically include quality control metrics, box plots, bar graphs and distribution plots of detected genes.
Advanced Biometrics analysis are used to provide additional insight into the sample and data obtained. The basic biometric analysis is typically expanded to include gene ontology; Differentially Expressed Gene (DEG) and pathway analysis.
Custom Biometrics involves the SeqLL team working with your laboratory in a partnering mode. This allows a combination of your molecule or target expertise with our analysis experience to gain non-intuitive insight into your data. Many projects involve re-evaluating existing data for additional correlation and enhanced discovery.


|